22 Oct
Taking microscopy imaging by STORM
Just over a decade ago, Xiaowei Zhuang and her team developed a novel microscopy imaging technique that changed the way we look at and study cells. STORM, or STochastic Optical-Reconstruction Microscopy has added significant momentum to the already-burgeoning field of super-resolution microscopy using various other methods including STED, pioneered by Stefan Hell and colleagues in Göttingen in mid-2000s. Now, over 10 years later, Xiaowei Zhuang has been awarded the 2019 Breakthrough Prize in Life Science for her ground-breaking research.
In the mid-2000s, super-resolution techniques were rapidly emerging as instrumental tools for many researchers working with cutting-edge microscopic imaging, and whose efforts of unravelling cellular microenvironments have been up until that point hindered by the seemingly insurmountable resolution limit of the optical systems they used. This meant that images of any objects positioned closer than approx. 200 nm coalesced into one blob in the final image. It was a major problem for anyone willing to study and resolve molecules at distances lower than that. Many fundamental biological processes occur at much smaller scales – single to low tens of nanometres - and having insight into this realm with a regular microscopy toolset would open entire new avenues of basic and applied research.
The STORM approach that the Zhuang lab took was different from STED insofar as it relied on a sequential switching of fluorophores on and off in a setup based on a wide field microscope. Based on sub-populations of these molecules emitting light at any given moment in time, it became possible to find out, or localize, where these light-emitting molecules were inside the cell or one of its many organelles. By taking repeated snapshots of various sub-populations, emitting light at different times and imaging them with an ultra-sensitive EMCCD camera, it became possible to create a full picture with individual positions of countless fluorophores resolved down to 20 nm.
It is clear why STORM and similar localisation techniques rely on efficient detection of single fluorophores. This can be challenging depending on the types of fluorophores used and, in some cases, such microscopic systems will need a camera detector of the highest sensitivity. EMCCD cameras have proved to be ideal imagers for non-point scanning super-resolution techniques. Their high quantum efficiency in excess of 95%, low noise and electron multiplication (EM) features all meant that even weaker fluorophores with lower photon yields could be efficiently detected by the camera. This, in turn, had a direct impact on the localisation precision and accuracy of the fluorophores and, consequently, biomolecules of interest.
Andor iXon EMCCDs have been at the forefront of this type of single molecule detection research since the early 2000s and have remained the cameras of choice for many types and variants of super-resolution microscopy. In their seminal 2006 paper, Xiaowei Zhuang and colleagues use a 512x512 pixels model of an iXon EMCCD (1). These cameras continue to be mainstay imagers for image-based super-resolution microscopy. The advent and continued refinement of scientific CMOS (sCMOS) cameras has seen this imaging technology make inroads into super-resolution microscopy and it is likely both technologies will coexist while addressing the different needs within various types of super-resolution imaging.
Applications of STORM are very varied and can now be seen across all research fields of bioimaging. They include studies of fluorophores’ characteristics (2), cytoskeletal rearrangements (3), receptor clustering (4), HIV infection (5), subcortical white matter (6) or lingual muscles function (7) with many more studies published with the help of Andor iXon EMCCDs every week.
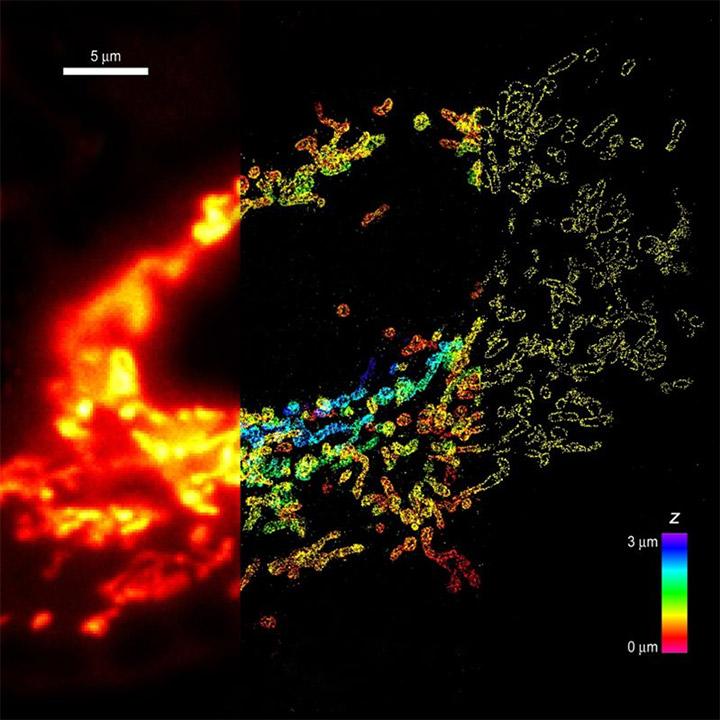
Whole-cell 3D STORM reveals interactions between cellular structures with nanometer-scale resolution - Comparison of conventional and 3D STORM image of mitochondria in a cell. The left panel shows the conventional image of the left 1/3 of the cell. The middle panel shows the 3D STORM image of the middle 1/3 of the cell. The z-dimension information is color-coded. The right panel shows the xy cross-section of the 3D STORM image of the right 1/3 of the cell. TOM20 in on mitochondria is immunolabeled with photoswitchable dyes. Imaging was done on Olympus IX71 equipped with Andor iXon 897 EMCCD.
From: B. Huang, S.A. Jones, B. Brandenburg, X. Zhuang; Nature Methods 5 1047-1052 (2008)
- Rust, M. J., Bates, M. & Zhuang, X. Nature Methods 3, 793–796 (2006)
- Dempsey GT et al., Nature Methods, 8(12), 1027–1036. (2011)
- Xu K., Babcock HP., Zhuang X., Nature Methods 9, 185–188 (2012)
- Rossboth et al., Nature Immunology 19, 821–827 (2018)
- Pereira CF et al., Virology Journal 9:84 (2012)
- Hainsworth AH et al., Neuropathology and Applied Neurobiology 44, 417–426 (2018)
- Cullins, MJ et al., The Laryngoscope (2017)

